Antimicrobial Resistance Genes in Pig Manure
This study examines environmental DNA (eDNA) from pig manure to map out and quantify antimicrobial resistance (AMR) genes. By using metagenome-assembled genomes (MAGs) and comparing eDNA sequences with known AMR gene databases, we reveal the types and abundances of bacteria and resistance genes in pig manure from three agricultural farms.
HIGHLIGHTS
- MAG Assembly: We employ a pipeline for the assembly of MAGs from metagenomic data, to describe the genetic content and potential function of bacteria within the microbial communities. This includes their capability for antibiotic resistance.
- Public Data and Open-Source Tools: The project leverages publicly available datasets and utilizes open-source software, such as Snakemake, to manage workflows. This approach underscores the accessibility and power of current bioinformatics tools in conducting high-quality genomic research.
- AMR Gene Profiling: Through targeted mapping and analysis, we quantify the abundance of AMR genes, offering insights into the spread and prevalence of antimicrobial resistance in pig manure across different farms.
THE PROJECT IN IMAGES
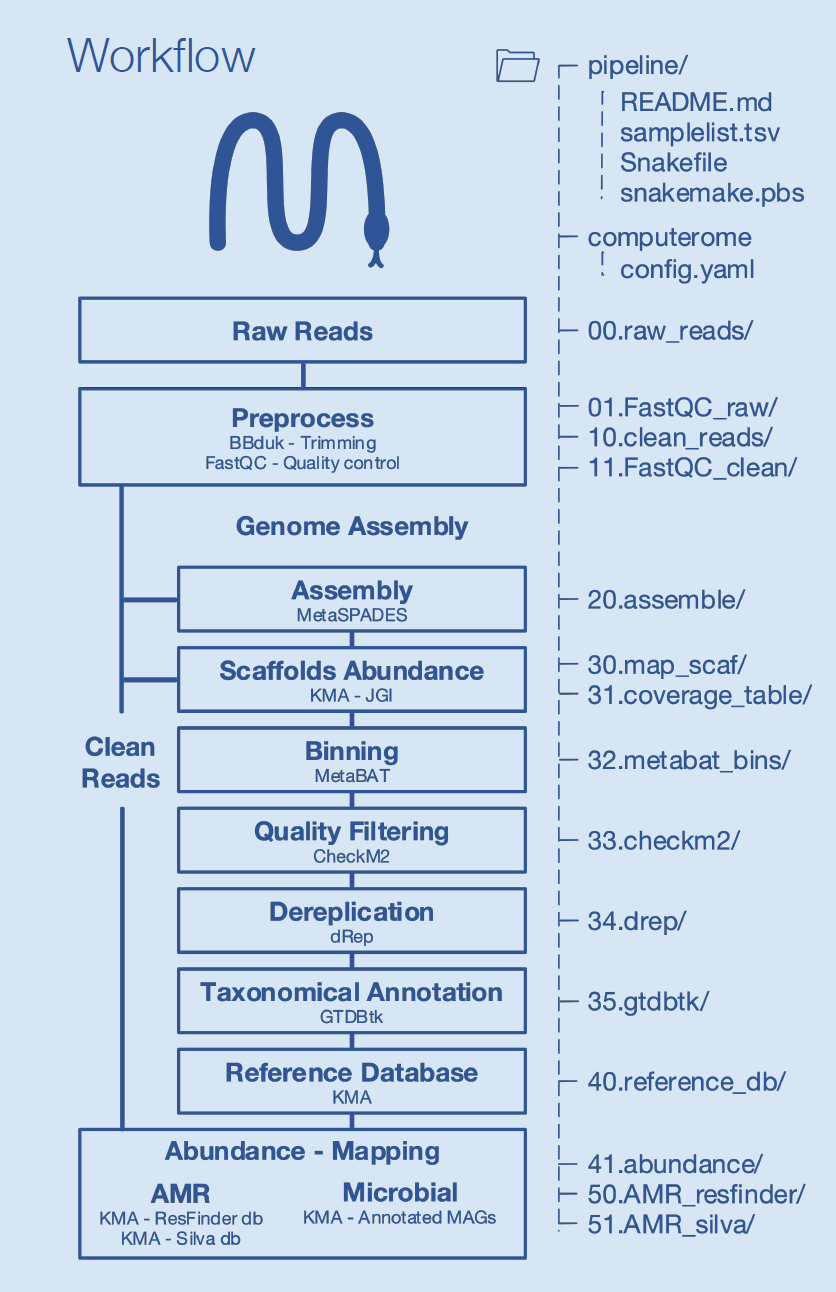
Figure 1: Overview of the metagenomics pipeline from raw reads through to taxonomical annotation and abundance mapping.
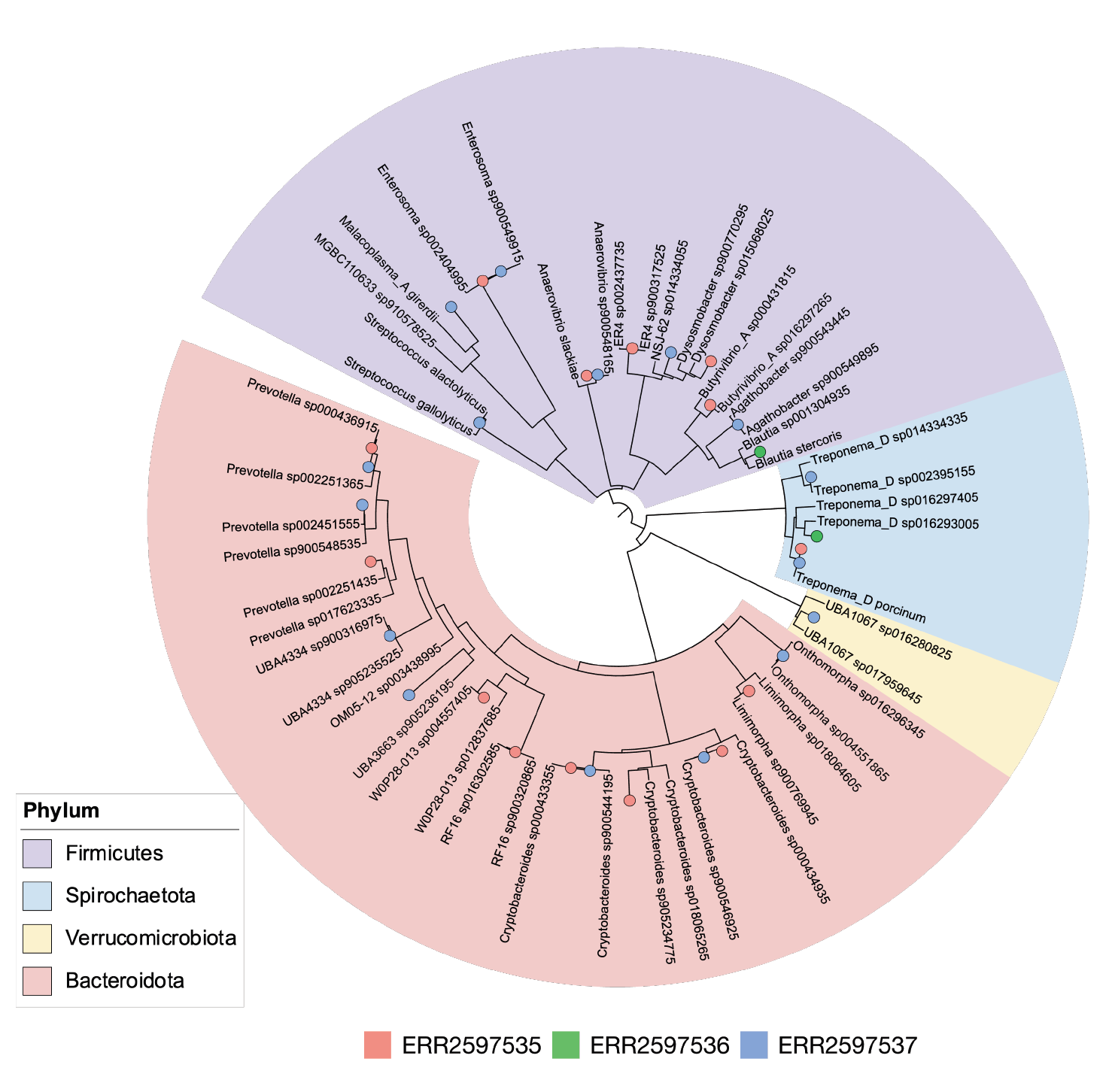
Figure 2: Phylogenetic tree showing the placement of metagenome-assembled genomes (MAGs) within various bacterial phyla for each of the three farms, ERR2597535, ERR2597536, ERR2597537.
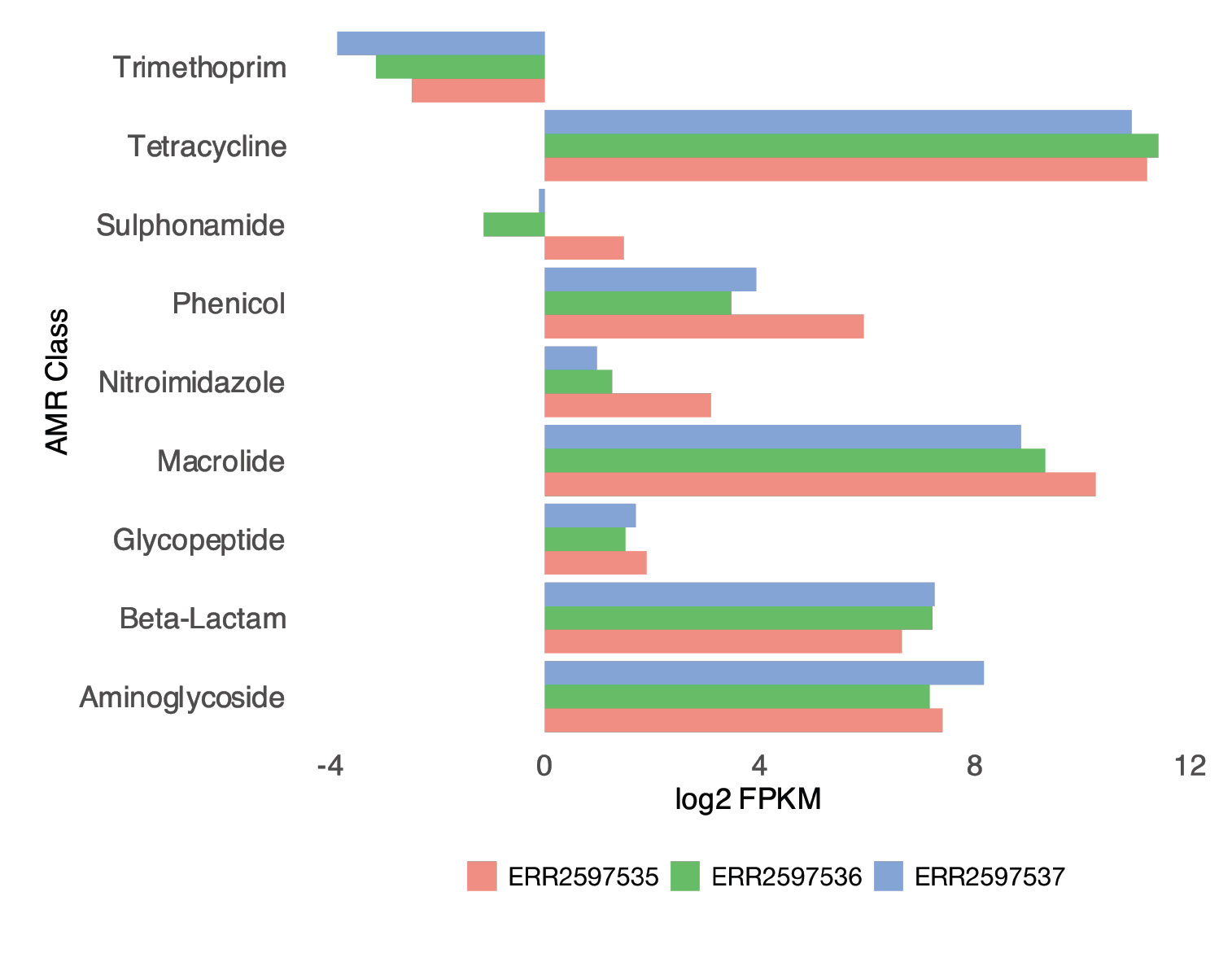
Figure 4: Relative abundance of antibiotic resistance gene classes, categorized by farm.
FURTHER INFORMATION
For details on our approach and results, please download our poster.